2621
Improving Lipid Suppression for 1H-MRSI Using Region-Optimized Virtual Coils1Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Cancer Center at Illinois, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States
Synopsis
One major challenge for 1H-MRSI with localized excitation is minimizing interference from undesired regions, particularly the subcutaneous lipids. In this work, we exploit the complementariness of a new technique called region-optimized virtual coils (ROVir) that is capable of generating a set of virtual channels sensitized to specific regions and spatiospectral processing for lipid removal. Using brain 1H-MRSI data, we demonstrated improved lipid removal by combining ROVir and a state-of-the-art union-of-subspaces method. We expect this integration to be useful for MRSI applications where VOIs are often lateralized (e.g., stroke and tumor) and/or cerebral lipids are of interest.
Introduction
One major challenge for localized MR spectroscopy and imaging is minimizing interference from undesired regions. An outstanding example is the subcutaneous lipids for 1H-MRSI, especially when the tissues of interest are close to the scalp. The typical selective excitation plus outer volume selection approach cannot flexibly adapt to the brain geometry1. Special hardware have been designed to null the lipid layers, e.g., using crusher coils2, RF shimming3, and/or high-order gradients for multidimensional RF designs4. However, the geometric definitions of the interference region for these approaches are still relatively simple (e.g., ellipsoids) and these instrumentations are highly customized and not easily accessible. Signal processing methods using different spatial-spectral constraints to better estimate and remove lipids have produced impressive results1,5,6, but performance still depends on the initial strength of the lipid signals. Region-optimized virtual coils (ROVir) is a new technique that generates a set of virtual coils (VCs) from the original arrays7, with maximized ratios between signals of interest from a predefined volume-of-interest (VOI) and signals from interference regions (referred to as SIR). In this work, we exploit the complementariness of ROVir and spatiospectral processing for lipid removal. Using brain 1H-MRSI data, we demonstrated improved lipid removal by combining ROVir and a state-of-the-art union-of-subspaces method. We expect this integration to be useful for MRSI applications where VOIs are often lateralized (e.g., stroke and tumor) and/or cerebral lipids are of interest.Methods
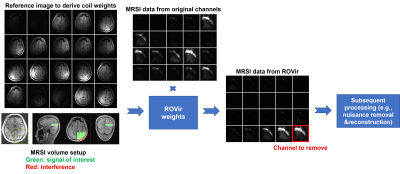
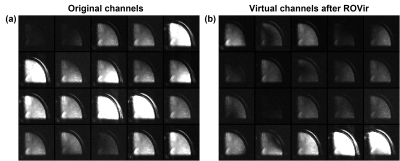
ROVir uses sensor-domain beamforming to generate a set of VCs that maximize the SIR between predefined VOI and interference region(s). The last VCs generated are typically localized to the interference region and can be removed to minimize its effects while protecting the VOI (see [7] for details). Here, we exploit this characteristic for improving lipid suppression in MRSI.MRSI data with volume selective excitation were acquired on a 3T scanner (Siemens Prisma) with IRB approval. Our experimental setup is illustrated in Fig. 1. Three ingredients are needed to use ROVir for lower-resolution MRSI data: (1) a multichannel high-resolution reference image (readily available in MRSI scans; we used MPRAGE here); (2) a mask for the VOI; and (3) a mask for the interference region (i.e., the lipid region). The masks can be obtained from segmentation of reference image and the aligned MRSI volume from GANNET8. As shown in Fig. 1, the original individual-channel MRS images (by spectral integration, water suppressed) are not uniquely sensitized to the undesired lipid region. After ROVir, the last VCs are more localized to the lipid region. Removing these channels can substantially reduce lipid contamination with minimal reduction of metabolite signals, which importantly is independent of resolution (a key difference to support-constrained k-space extrapolation). Afterwards, the remaining channels can be combined for additional nuisance removal and spatiospectral reconstruction6,9. As we expect ROVir to be most effective when the volume-of-excitation is lateralized to one side of the brain (due to coil configuration), the data shown below were from excitation volumes chosen in this fashion (see Fig. 1, bottom of first column). This volume selection, mimicking a situation where MRSI of a lateralized lesion is needed, inevitably included the subcutaneous lipid layer.
Results
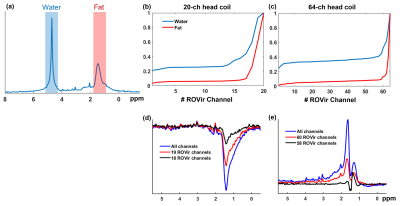
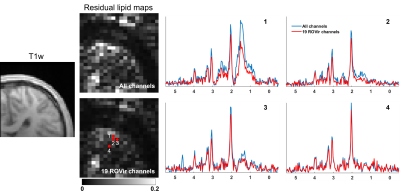
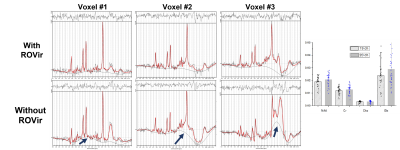
Figure 2 compares the signals from water and lipid peaks w.r.t. different numbers of ROVir channels kept for an in vivo CSI data. The data were acquired using a semi-LASER sequence with: volume-of-excitation=65x65x10mm3 (location described above), FOV=100x100mm2, matrix size=24x24, TR/TE=1200/50ms and spectral BW=2000Hz. For the 20-channel coil, removing the last ROVir channel reduced the lipid signals by nearly 2X with only mild loss for water. For the 64-channel coil, we can remove the last 4 ROVir channels for effective lipid reduction with small signal loss, evident in both the tradeoff curve (Fig.2b-c) and the localized spectra (Fig.2d-e, water removed). We chose water peak because the metabolite peaks can be contaminated by the tails of the strong lipid peaks and bias the assessment. High-resolution nonwater-suppressed sLASER EPSI scans were also acquired from the same volume as the CSI to better assess the characteristics of the ROVir channels (Fig. 3).To evaluate the complementary power of ROVir and advanced signal processing strategies for lipid removal, we applied a union-of-subspaces model-based nuisance removal to the ROVir processed data. We call this overall strategy ROVirSpect. Figure 4 compares the overall removal performance for ROVSpect and a direct removal without ROVir for a 20-ch CSI dataset. ROVSpect substantially reduced the residual lipid leakage into the brain, with stronger effects for the edge voxels. The metabolite peaks were only slightly affected which further supported the utility of ROVirSpect. For a more thorough analysis, we also performed a B0-inhomogeneity corrected reconstruction9 of the data and metabolite quantification of the reconstructions using LCModel10. Spectra from ROVirSpect lead to statistically different fits and less variable baseline compared to results produced without ROVir (Fig. 5).
Discussion and Summary
The combination of ROVir and spatiospectral constrained nuisance removal improved lipid suppression for 1H-MRSI with volume selective excitation that contains subcutaneous layers. The improved lipid suppression was achieved with only mildly affected metabolite components. We note that the performance of ROVirSpect is dependent on the coil configuration and the location of the excitation volume, which should be further evaluated for practical applications.Acknowledgements
This work was supported in part by NSF-CBET-1944249, NIH-NIBIB-1R21EB029076A and a seed grant from the Cancer Center at the University of Illinois at Urbana-Champaign.References
[1] Wilson M et al. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn Reson Med. 2019;82:527-550.
[2] Boer VO et al. Lipid suppression for brain MRI and MRSI by means of a dedicated crusher coil. Magn Reson Med. 2015;73:2062-2068.
[3] Hetherington HP et al. RF shimming for spectroscopic localization in the human brain at 7 T. Magn Reson Med. 2010;63:9-19.
[4] de Graaf RA et al. Elliptical localization with pulsed second-order fields (ECLIPSE) for robust lipid suppression in proton MRSI. NMR Biomed. 2018;31:3949.
[5] Bilgic B et al. Lipid suppression in CSI with spatial priors and highly undersampled peripheral k-space. Magn Reson Med. 2013;69:1501-1511.
[6] Ma C et al. Removal of nuisance signals from limited and sparse 1H MRSI data using a union-of-subspaces model. Magn Reson Med. 2016;75:488-497.
[7] Kim D et al. Region-optimized virtual (ROVir) coils: Localization and/or suppression of spatial regions using sensor-domain beamforming. Magn Reson Med. 2021;86:197-212.
[8] Edden RAE et al. GANNET: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40:1445-1452.
[9] Lam F et al. High‐resolution 1H‐MRSI of the brain using SPICE: Data acquisition and image reconstruction. Magn Reson Med. 2016;76:1059-1070.
[10] Provencher SW et al. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med. 1993;30:672–679.
Figures