1014
Improving the Sensitivity to Myocardial Scar in Automatic Segmentations of Left Ventricular Myocardium on LGE Images
Marc Vornehm1,2, Maximilian Fenski3, Elisabeth Preuhs4, Andreas Maier4, Jeanette Schulz-Menger3, Jens Wetzl1, and Daniel Giese1,5
1Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany, 2Artificial Intelligence in Biomedical Engineering, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 3Working Group Cardiovascular Magnetic Resonance, Experimental and Clinical Research Center, Charité Medical Faculty, Max Delbrück Center for Molecular Medicine, HELIOS Klinikum Berlin Buch, Department of Cardiology and Nephrology, Berlin, Germany, 4Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 5Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany
1Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany, 2Artificial Intelligence in Biomedical Engineering, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 3Working Group Cardiovascular Magnetic Resonance, Experimental and Clinical Research Center, Charité Medical Faculty, Max Delbrück Center for Molecular Medicine, HELIOS Klinikum Berlin Buch, Department of Cardiology and Nephrology, Berlin, Germany, 4Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 5Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany
Synopsis
Automatic segmentation of left ventricular myocardium on LGE images of patients with chronic myocardial infarction is challenging due to inhomogeneous signal intensities in the presence of myocardial scar. However, the inclusion of scar in automatically generated myocardium segmentations is critical for further LGE analysis. We propose a Deep Learning-based method for myocardial segmentation on cardiac LGE images, achieving average Dice scores of 0.78 and 0.80 on test images with and without scar, respectively. We furthermore evaluate the network’s sensitivity to scar and show that it can be improved by incorporating synthetic images with diverse enhancement patterns in the training data.
Introduction
Contouring of the left ventricular (LV) myocardium in late gadolinium enhancement (LGE) images is an essential step in quantification of myocardial scar and is usually performed manually by the reader1. Automatic segmentation of the LV could facilitate this process and could furthermore serve as starting point for a subsequent fully automatic quantification of myocardial scar2. However, automatic segmentation on LGE images is challenging due to highly variable enhancement patterns, inhomogeneous image intensities within the scarred region, and ambiguous image boundaries, particularly in patients with subendocardial scar. Inclusion of scarred tissue within the myocardial boundaries, however, is critical for an accurate scar quantification in the follow-up. Multiple Deep Learning-based methods for myocardial segmentation have been proposed previously, achieving Dice similarity coefficients (DSC) of around 0.78-0.823-7. However, they were not quantitatively evaluated for their sensitivity to myocardial scar, i.e., the percentage of scar included in the myocardial segmentation. In this study, we describe a U-Net-based approach for myocardial segmentation and evaluate its performance in scarred regions. We investigate the effect of incorporating simulated images, hypothesizing that an increased variability in enhancement patterns in the training data improves the network’s sensitivity to scar.Methods
Fifty-one patients with chronic myocardial infarction underwent motion corrected PSIR-LGE imaging on a 1.5T scanner (MAGNETOM AvantoFit, Siemens Healthcare, Erlangen, Germany) ~15-20 minutes after injection of 0.15mmol/kg Gadoteridol (slice thickness 6mm, resolution 1.56mm²). Each acquisition was reconstructed as magnitude inversion recovery (MagIR) and phase-sensitive inversion recovery (PSIR)8. Myocardial borders were contoured by a reader with three years of experience (consensus read with an SCMR Level III certified reader in case of inconclusive findings). Corresponding cine images were used as reference to support the contouring process. Enhanced regions were observed in 44 patients (LGE-positive) and the scar area was determined using the full width at half maximum (FWHM) method9,10.The data set was supplemented with synthetic images created using a Bloch simulation framework and the XCAT phantom11. Scars were simulated by altering the tissue parameters within the myocardium. The simulations included variations in anatomy, scar location, size, and transmurality, and parameters influencing the image contrast. 300 acquisitions were simulated and reconstructed as MagIR and PSIR images. Exemplary real and simulated images are given in Figure 1.
Images were cropped to 256x256 pixels with the heart centered. Of the 44 LGE-positive patients, 15 were used for training and five for validation. The remaining 24 LGE-positive patients and seven LGE-negative patients (i.e., without myocardial scar) were set aside and formed two separate data sets for testing. MagIR and PSIR images were used concurrently, resulting in 30 individual images for training. The number of synthetic images in the training set was varied between zero and 600. Images were augmented using affine transformations, blurring, and local image deformation at a random location within the myocardium. The real images were presented to the network twenty times per training epoch in randomly augmented ways, such that real images were presented to the network at least as often as synthetic images. The network was based on the U-Net architecture12 with residual connections, BatchNormalization and ReLU activation functions (Figure 2). It was optimized using the Dice loss function13 and Adam optimizer14 with a learning rate of 10-3. The obtained segmentation masks were post-processed by removing all but the largest connected component.
Results
The trained networks were evaluated in terms of average DSC and sensitivity to myocardial scar. Average DSC was higher on the LGE-negative (0.79-0.80) than on the LGE-positive test set (0.76-0.78) in all experiments. Adding synthetic data did not significantly affect the DSC. The average sensitivity to myocardial scar was evaluated on the LGE-positive test set and was observed to increase when more synthetic images were added to the training set. Detailed quantitative results and exemplary segmentation results of images with myocardial scar are given in Figures 3 and 4, respectively.Discussion
The reported DSC values are comparable to those of previously published methods and were achieved using a standard network architecture and very few training samples. A more accurate segmentation of the myocardium on LGE images is mainly hindered by the presence of enhanced regions, i.e., scarred tissue. This is manifested in the decreased DSC observed in LGE-positive images compared to LGE-negative images. The reported sensitivity values for myocardial scar indicate that considerable portions of the scars were not included in the myocardial borders. Subendocardial scars, exhibiting similar signal intensities to the adjacent blood pool, are particularly prone to exclusion from automatic segmentations (Figure 4d). Increasing the number of synthetic images in the training set led to improved sensitivity values, possibly due to an increased variability of enhancement patterns in the training data.Conclusion
We presented an automatic approach for myocardial segmentation on LGE images, achieving similar results as previous methods in terms of DSC. The clinical value of such algorithms, however, is considerably diminished if the sensitivity to myocardial scar is low. We showed that this issue can be addressed by increasing the variability of enhancement patterns in the training data, specifically by incorporating simulated images. Other approaches like, for instance, style transfer methods for image synthesis6 or innovative augmentation techniques5 could similarly be of interest in this regard.Acknowledgements
No acknowledgement found.References
- Schulz-Menger J, Bluemke D, Bremerich J, et al. Standardized image interpretation and post-processing in cardiovascular magnetic resonance - 2020 update: Society for Cardiovascular Magnetic Resonance (SCMR): Board of Trustees Task Force on Standardized Post-Processing. J Cardiovasc Magn Reson. 2020;22:19.
- Moccia S, Banali R, Martini C, et al. Development and testing of a deep learning-based strategy for scar segmentation on CMR-LGE images. Magn Reson Mater Phy. 2019;32(2):187-195.
- Fahmy AS, Rausch J, Neisius U, et al. Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. JACC-Cardiovasc Imag. 2018;11(12):1917-1918.
- Yue Q, Luo X, Ye Q, et al. Cardiac Segmentation from LGE MRI Using Deep Neural Network Incorporating Shape and Spatial Priors. In: Shen D, et al. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. 2019;559-567.
- Campello VM, Martín-Isla C, Izquierdo C, et al. Combining Multi-Sequence and Synthetic Images for Improved Segmentation of Late Gadolinium Enhancement Cardiac MRI. In: Pop M, et al. (eds) Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges. STACOM 2019. 2020;290-299.
- Chen C, Ouyang C, Tarroni C, et al. Unsupervised Multi-modal Style Transfer for Cardiac MR Segmentation. In: Pop M, et al. (eds) Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges. STACOM 2019. 2020;209-219.
- Roth H, Zhu W, Yang D, et al. Cardiac Segmentation of LGE MRI with Noisy Labels. In: Pop M, et al. (eds) Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges. STACOM 2019. 2020;228-236.
- Kellman P, Arai A, McVeigh E, et al. Phase-sensitive inversion recovery for detecting myocardial infarction using gadolinium-delayed hyperenhancement. Magn Reson Med. 2002;47(2):372-383.
- Amado L, Gerber B, Gupta S, et al. Accurate and objective infarct sizing by contrast-enhanced magnetic resonance imaging in a canine myocardial infarction model. J Am Coll Cardiol. 2004;44(12):2383-2389.
- Flett A, Hasleton J, Cook C, et al. Evaluation of Techniques for the Quantification of Myocardial Scar of Differing Etiology Using Cardiac Magnetic Resonance. JACC-Cardiovasc Imag. 2011;4(2):150-156.
- Segars WP, Sturgeon G, Mendonca S, et al. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010;37(9):4902–4915.
- Ronneberger O, Fischer P, Brox T, et al. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, et al. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. 2015;234-241.
- Milletari F, Nassir N, Seyed-Ahmad A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV). 2016;565-571.
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv:1412.6980 [cs.LG]. 2017.
Figures
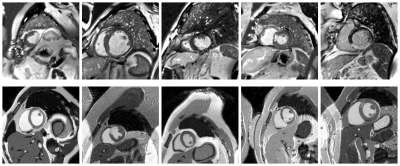
Figure 1: Exemplary LGE images with
myocardial scar (Top: Real; Bottom: Simulated). Enhancement patterns observed
in LGE images vary in transmurality, location, and size.
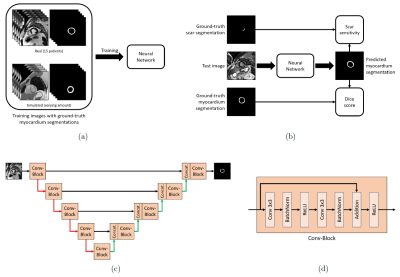
Figure 2: (a) The network is trained using real
and simulated images with corresponding ground-truth myocardium masks. (b) The
network is tested using previously unseen data. (c) The network architecture is
based on the U-Net. Red arrows indicate max-pooling, green arrows indicate up-sampling
followed by a convolutional layer, black arrows indicate concatenation. (d) Internal
structure of each Conv-Block indicated in (c).
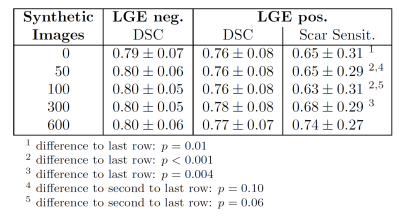
Figure 3: Detailed results in terms of
average Dice score (DSC) and sensitivity to myocardial scar pixels for two
different test data sets (LGE negative = without scar, LGE positive = with scar).
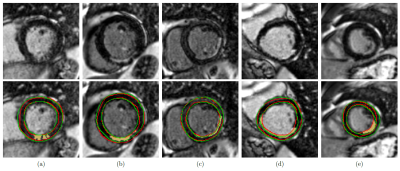
Figure 4: Close-ups on exemplary
segmentation results. Top row: Original images. Bottom row: Corresponding
segmentations (red = ground-truth myocardium contour, green = myocardium
contour obtained by our method, yellow = ground-truth scar mask). (a)-(b)
transmural scars fully included in the obtained myocardium mask. (c)
subendocardial scar partially included in the myocardium mask. (d)
subendocardial scar not included in the myocardium mask. (e) transmural scar
not included in the myocardium mask.
DOI: https://doi.org/10.58530/2022/1014