3693
Whole-brain B1-corrected quantitative MT imaging in less than 5 minutes1Department of Radiology, Division of Radiological Physics, University Hospital Basel, Basel, Switzerland, 2Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 3Department of High Field Magnetic Resonance, Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 4Department of Biomedical Magnetic Resonance, University of Tübingen, Tübingen, Germany, 5Department of Biomedical Engineering, University of Virginia, Virginia, VA, United States, 6Department of Application Development, Siemens Healthcare, Erlangen, Germany
Synopsis
To develop a rapid quantitative magnetization transfer (qMT) imaging methodology that offers whole brain coverage and takes into account B1-field inhomogeneities for improved parameter estimation accuracy in the clinical setting.
Introduction
Quantitative magnetization transfer (qMT) imaging has shown potential for the prognosis and diagnosis of different neurological abnormalities, such as multiple sclerosis (1). Estimation of qMT tissue parameters from a two-pool model system, however, is currently limited in the clinical setting by long data acquisition times (2). Recently, for rapid whole-brain magnetization transfer ratio (MTR) imaging, the use of a prototype multi-slice MT-weighted spoiled gradient echo (SPGR) sequence was suggested, offering whole brain MTR imaging with intrinsic B1-correction within less than one minute (3). The method takes advantage of a rapid interleaved spiral readout offering whole-brain coverage for the acquisition of a single MT-weighted volume within 20 seconds. The same underlying spiral SPGR prototype was also used for rapid B1-corrected whole-brain T1 mapping, again in less than one minute (4). In this work, we propose to fuse both concepts for rapid in-vivo qMT parameter estimation from a two-pool model system. We will show that the spiral prototype offers whole-brain B1-corrected quantitative estimation of all qMT tissue parameters within less than 5 minutes.Methods
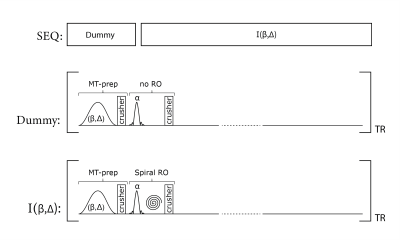
MT-weighted whole-brain volumes are acquired using a rapid multi-slice SPGR prototype sequence, as described in (3,4). In this work, a single high-resolution MT-weighted volume is reconstructed from the acquisition of 50 interleaved slices with a TR of 1650 ms. For each acquisition, the sequence kernel (effective TR of 33 ms) consisted of a MT-preparation module (with gaussian-shaped saturation flip angle β, offset frequency Δ, and duration of 17.92 ms) followed by the slice excitation (3mm thickness, sinc-shaped RF pulse of 0.6 ms duration, and flip angle of α=25°) using an interleaved spiral readout (14.2ms duration). For each slice, 20 interleaved spiral trajectories were acquired and reconstructed to a 1.3 x 1.3 mm2 in-plane resolution using a spiral version of the iterative self-consistent parallel imaging reconstruction method (SPIRiT) in combination with parallel imaging of factor two (5). From the multi-slice acquisition, the repetitive excitation, and the interleaved spiral readout, the spin magnetization of a single slice effectively preceded a pulse train of 50 MT-saturation pulses. The overall acquisition time for a single MT-weighted whole-brain volume was 20 seconds (including a dummy preparation period of 4s without readout but with MT-preparation and excitation pulses to put the two-pool system in the steady-state). For an overview, see Figure 1.In addition, a B1-field and an observable T1-map (T1,obs) were acquired based on the same spiral prototype sequence, described in (3). Essentially, the same resolution was used as for the MT-weighted scans described before. The overall acquisition time for the T1-B1 mapping was 53 seconds.
Two-pool model parameter estimation was performed from a set of 12 MT-weighted volumes in combination with the T1,obs and B1 map as described in (6). To this end, a super-Lorentzian line-shape was used and, since the model is not sensitive to the longitudinal relaxation rate of the bound pool fraction (R1r), it was further assumed that R1r = R1f = 1/T1,obs. The set of MT-weighted volumes were acquired with Δ = 1kHz, β = 150°,250°,350°,450°,550°,650°,750°,850° and Δ = 6kHz, β = 250°,450°,650°,850°. The MT flip angle was modulated by the corresponding B1-field prior to the estimation of remaining two-pool model parameters, that is the bound proton fraction (F), the exchange rate (kf), and the transverse relaxation time of the bound pool protons (T2r).
Three volunteers were scanned at 3T (Magnetom Prisma, Siemens Healthcare, Erlangen, Germany) using a 20-channel receive head coil. Measurements were approved by our local ethics committee. Image postprocessing was conducted by MATLAB R2019a (The MathWorks, Inc., Natick, MA). The standard software package FSL (FMRIB Software Library v6.0, Oxford, UK) was used for co-registration of images. Whole-brain imaging of 12 MT-weighted images together with T1-B1 mapping was completed within 4 minutes and 53 seconds.
Results
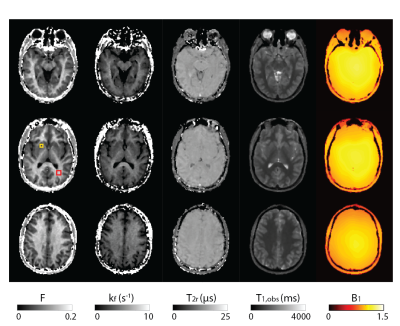
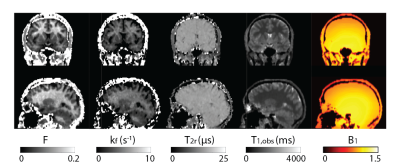
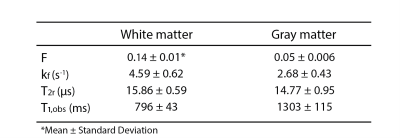
Figure 2 shows all the retrieved quantitative parameter estimation for three exemplary axial slices, that is F, kf, T2r, T1,obs, and B1. Coronal and sagittal views of the results are presented in Figure 3. For a selected region-of-interest (ROI) in white and gray matter (cf. Figure 2), parameter estimations are given in Table 1. Overall, the retrieved two-pool model parameters are in good agreement with the literature (7).Discussion
Recently, a spiral prototype sequence was proposed for rapid whole-brain B1-corrected T1 and MTR quantification in less than one minute. In this work, we developed this approach further to offer rapid whole-brain qMT imaging in clinically relevant acquisition times. The T1-B1 mapping method yields a simultaneous B1-map that was used not only to remove the bias in the T1 estimate but also to improve the overall accuracy of the two-pool model parameter estimation. In this work, we targeted robust whole-brain qMT imaging in clinically relevant acquisition times, within 5 minutes. To this end, sampling of the MT-response was confined to 12 acquisitions with two different frequencies and a variety of different MT flip angles.Conclusion
In conclusion, we have shown that whole-brain B1-corrected qMT imaging is feasible within less than 5 minutes with clinically relevant resolutions at 3T using an interleaved spiral prototype sequence. The proposed approach thus offers excellent prospects for wide clinical translation and use.Acknowledgements
This work was supported by the Swiss National Science Foundation (SNF grant No. 325230_182008).References
1. Paul Tofts. Quantitative MRI of the Brain: Measuring Changes Caused by Disease. 2004. 650 p.
2. Cercignani M, Dowell NG, Tofts PS. Quantitative MRI of the Brain: Principles of Physical Measurement, Second edition. CRC Press; 2018. 361 p.
3. Heule R, Pfeuffer J, Meyer CH, Bieri O. Simultaneous B1 and T1 mapping using spiral multislice variable flip angle acquisitions for whole-brain coverage in less than one minute. Magn Reson Med. 2019;81(3):1876–89.
4. Afshari R, Santini F, Heule R, Meyer CH, Pfeuffer J, Bieri O. One-minute whole-brain magnetization transfer ratio imaging with intrinsic B1-correction. Magn Reson Med. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.28618
5. Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med. 2010;64(2):457–471.
6. Sled JG, Pike GB. Quantitative interpretation of magnetization transfer in spoiled gradient echo MRI sequences. J Magn Reson. 2000 Jul;145(1):24–36.
7. Sled JG, Pike GB. Quantitative imaging of magnetization transfer exchange and relaxation properties in vivo using MRI. Magn Reson Med. 2001;46(5):923–31.
Figures