2714
The Open Source Initiative for Perfusion Imaging (OSIPI) ASL MRI Challenge
Udunna Anazodo1,2, Joana Pinto3, Flora A. Kennedy McConnell4,5,6, Maria-Eleni Dounavi7, Cassandra Gould van Praag8, Henk Mutsaerts9, Aaron Oliver Taylor10, André Paschoal11, Jan Petr12, Diego Pineda-Ordóñez13, Joseph G. Woods14, Moss Y. Zhao15, and Paula L. Croal4,5
1Department of Medical Biophysics, University of Western Ontario, London, ON, Canada, 2Imaging Division, Lawson Health Research Institute, London, ON, Canada, 3Institute of Biomedical Engineering, Department of Engineering Science, University of Oxford, Oxford, United Kingdom, 4Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, United Kingdom, 5Radiological Sciences, Division of Clinical Neuroscience, School of Medicine, University of Nottingham, Nottingham, United Kingdom, 6Nottingham Biomedical Research Centre, Queens Medical Centre, Nottingham, United Kingdom, 7Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 8Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 9Amsterdam University Medical Center, Amsterdam, Netherlands, 10Gold Standard Phantoms Limited, London, United Kingdom, 11Department of Radiology, LIM44 - HCFMUSP, Sao Paulo, Brazil, 12Helmholtz-Zentrum Dresden-Rossendorf, Dresden, Germany, 13Department of Radiology, Clinica Del Country, Bogotá, Colombia, 14Department of Radiology, University of California San Diego, San Diego, CA, United States, 15Department of Radiology, Stanford University, Stanford, CA, United States
1Department of Medical Biophysics, University of Western Ontario, London, ON, Canada, 2Imaging Division, Lawson Health Research Institute, London, ON, Canada, 3Institute of Biomedical Engineering, Department of Engineering Science, University of Oxford, Oxford, United Kingdom, 4Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, United Kingdom, 5Radiological Sciences, Division of Clinical Neuroscience, School of Medicine, University of Nottingham, Nottingham, United Kingdom, 6Nottingham Biomedical Research Centre, Queens Medical Centre, Nottingham, United Kingdom, 7Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 8Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 9Amsterdam University Medical Center, Amsterdam, Netherlands, 10Gold Standard Phantoms Limited, London, United Kingdom, 11Department of Radiology, LIM44 - HCFMUSP, Sao Paulo, Brazil, 12Helmholtz-Zentrum Dresden-Rossendorf, Dresden, Germany, 13Department of Radiology, Clinica Del Country, Bogotá, Colombia, 14Department of Radiology, University of California San Diego, San Diego, CA, United States, 15Department of Radiology, Stanford University, Stanford, CA, United States
Synopsis
The OSIPI ASL MRI Challenge is a community-led initiative aiming to establish the range of approaches used for ASL image analysis and cerebral blood flow (CBF) quantification. Challenge data will consist of population-based and synthetic pseudo-continuous ASL images, with participants analysing the data and submitting resulting CBF maps and mean tissue CBF, along with documentation. Entries will be scored on accuracy, reproducibility and documentation quality. Through documenting the analysis choices made within the community, we will begin to better understand sources of variability, ultimately identifying an optimum pipeline, and moving towards the much-needed consensus of ASL image processing standards.
Introduction
There is an established record of MRI focused challenges1-4, which draw on community expertise to validate novel methods, as well as characterise and optimise current practices. Given the significant movement towards clinical translation of arterial spin labeling (ASL) MRI, but lack of community consensus to image processing and quantification of perfusion parameters, the ASL challenge proposed here is timely.There has been an initial consensus for clinical application of ASL MRI5, focussed mainly on acquisition. Consensus on post-processing is equally important, as well as understanding sources of variability and bias introduced by different analysis approaches. Despite efforts to develop ASL image processing standards6-11, community-wide consensus remains to be established.
Here, we introduce a challenge focused on image analysis and quantification of ASL cerebral blood flow (CBF) maps, as part of the ISMRM perfusion workgroup Open Source Initiative for Perfusion Imaging (OSIPI)12.
Aims
Our main objectives are to compare existing post-processing pipelines (freely available, in-house and/or commercial) for image analysis and quantification of ASL-measured cerebral perfusion, identify optimum approaches, and document the variety of post-processing tactics used for ASL MRI across the community.Challenge Overview
Structure: Challenge design and accompanying information will be accessible on the ISMRM challenge website13. Following online registration, challenge data will be made available via an online repository. The challenge will be open for entries for six months from launch (February 2021, Figure 1).Data: Challenge datasets will consist of ten pseudo-continuous ASL (PCASL) datasets in ASL BIDS14 format: 4D timeseries in control-label format, with a separate M0 image, and high-resolution T1-weighted anatomical image, from two sources (Figure 2):
(i) Population-based Data: An existing population-based Digital Reference Object (DRO)11 will be used to establish variability across ‘real-world’ data. Data consists of 3T PCASL (PLD: 2025-3310ms, 2D EPI, TE/TR=10.4/4800ms, 1650ms labeling duration, 3.5x3.5x4.5 mm3 resolution, 30 control-label pairs), and 3D TFE (TE/TR=3.14/6.76ms, FA=9°, 1mm isotropic resolution) with M0 and control-label pairs averaged across 84 elderly participants (67.1±7.1 years)15.
(ii) Synthetic Data: Nine datasets will be simulated using an existing ASL-DRO16. Ground-truth datasets were simulated in subject-specific anatomical space, using 3D T1-weighted images from healthy young adults17 (MPRAGE, TE/TR=2.14/2400ms, FA=8°, 0.7mm isotropic resolution) and subsequently downsampled to ASL resolution (4x4x4 mm3). Input parameters (CBF, arterial transit time, M0, T1, T2 and T2*) were defined per tissue type (grey matter/white matter/cerebrospinal fluid, GM/WM/CSF) for simulation of ASL signal according to the General Kinetic model18 and 3D PCASL timeseries acquired at 3T, with gradient-echo readout, consistent with consensus recommendations5 (PLD: 1800ms, TE/TR: 10.4/4800ms, 1800ms labelling duration, M0 TR=10s, 30 control-label pairs). For challenge purposes, simulation parameters will be withheld. Simulations are to reflect both normality and pathology (modulated regional perfusion and/or tissue ratios), with motion levels manipulated across a realistic range of motion. Specific modulation details will be limited to a maximum of three investigators (UA, JP and MZ).
Submission: For each dataset, participants will be required to submit a CBF map in ASL native space, along with mean CBF values for GM/WM regions, and accompanying tissue segmentations. If partial volume correction has been performed, participants will be asked to also submit uncorrected results, and additional tissue segmentations, if used. Documentation (maximum 3 pages) should provide sufficient detail for replication of analysis. Documentation requirements will be provided in line with COBIDAS recommendations19. Participants may use in-house pipelines, or re-use existing pipelines in part or in whole. For in-house pipelines, accompanying code is encouraged for reproducibility purposes. Should commercial software be used, participants must be able to share a temporary license for scoring purposes.
Scoring: Entries will be scored out of 100 for (i) accuracy, (ii) reproducibility and (iii) documentation quality, combined into a single final score with 70:20:10 weighting, respectively. Accuracy will be assessed through quantitative comparison to ground-truth for both voxel-wise and region-wise perfusion measures. Submissions will be reproduced by the challenge organisers according to submitted documentation (and code where appropriate), with voxelwise and mean tissue perfusion comparisons made between submitted and reproduced data. Documentation will be scored on quality20 and on how many minimum requirements are met. Population-based results will establish real-world variability within the community, but will not contribute to quantitative scoring due to the lack of ground-truth. Scores will be available online within six months after challenge close, with the three highest scoring participants invited to present at the annual ISMRM perfusion study group meeting.
Discussion
The OSIPI ASL MRI Challenge aims to establish an overview of the variety of ASL image analysis and quantification choices made by the community for common datasets. By emphasising the importance of methods documentation, we aim to encourage good scientific practice, increasing reproducibility of results. By encouraging both developers and general users to take part we aim to encompass a broad range of backgrounds reflecting the diverse nature of the ASL community. Finally, this challenge may lead to understanding sources of variability and may ultimately identify an optimum ASL pipeline.Acknowledgements
HM is supported by the Dutch Heart Foundation (2020T049), and by the Eurostars-2 joint programme with co-funding from the European Union Horizon 2020 research and innovation programme, provided by the Netherlands Enterprise Agency (RvO). PLC is supported by The Brain Tumour Charity.References
- Pujol et al, The DTI challenge: toward standardized evaluation of diffusion tensor imaging tractography for neurosurgery. J Neuroimaging, 2015; 25(6):8782
- https://www.nitrc.org/projects/nrm2018_petgc
- Grissom et al., Advancing RF pulse design using an open-competition format: report from the 2015 ISMRM challenge. 2017; 78(4): 1352-1361
- Menze et al., The multimodal brain tumour image segmentation benchmark (BRATS). IEEE Trans Med. Imaging, 2015; 34(10):1993-2004
- Alsop et al., Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. MRM, 2015; 73(1):102-16
- Wang et al., Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx, MRM., 2008; 26(2):262-269
- Li et al., ASL-MRICloud: an online tool for the processing of ASL MRI data, NMR Biomed., 2019; 32(2), e4051
- Chappell et al., Variational Bayesian inference for a non-linear forward model. IEEE Transactions on Signal Processing., 2009: 57(1):223-236
- Mato Abad et al., ASAP (automatic software for ASL processing): a toolbox for processing arterial spin labeling images, MRM., 2016; 34(3):334-344
- Almeida et al., Test-retest reliability of cerebral blood flow in healthy individuals using arterial spin labeling: findings from the EMBARC study, MRM., 2018; 45(2018):26-33
- Mutsaerts et al., ExploreASL: An image processing pipeline for multi-center ASL perfusion MRI studies. NeuroImage, 2020; 219:117031
- www.osipi.org
- https://challenge.ismrm.org/
- Clement et al., ASL-BIDS, the brain imaging data structure extension for arterial spin labeling. Magn. Reson. Mat. Phys. Biol. Med. 2019; 32(1 Supplement)
- Ritchie et al., Development of intervention for the secondary prevention :117031of alzheimer’s Dementia (EPAD) project. Lan. Psychiatr. 2016; 3(2):179-186
- Pypi.org/project/asldro
- Van Essen et al., The WU-Minn Human Connectome Project: An Overview. NeuroImage, 2013; 80:62-79
- Buxton et al., A general kinetic model for quantitative perfusion imaging with arterial spin labelling. MRM, 1998; 40(3):383-396vol. 40, no. 3, pp. 383–396, Sep. 1998
- Nichols et al., Best practices in data analysis and sharing in neuroimaging using MRI, Nat. Neurosci. 2017; 20(3):299-303
- O’Keefe, 2010, Accessed from https://www.scriptorium.com/2010/05/calculating-document-quality-quack/
Figures
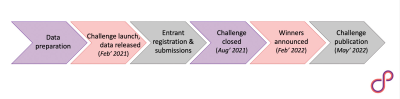
Figure 1: Projected timeline for the OSIPI ASL Challenge. The challenge will be open for a period of six months, with winning teams presenting at the ISMRM Perfusion Study Group meeting at ISMRM 2022.
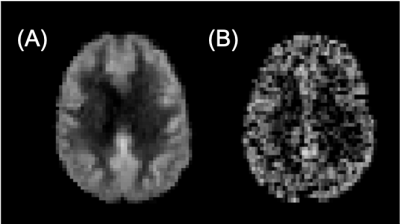
Figure 2: Example perfusion-weighted images (arbitrary units) for A) population-based and B) synthetic datasets.