2417
Prospective Performance Evaluation of the Deep Learning Reconstruction Method at 1.5T: A Multi-Anatomy and Multi-Reader Study1Canon Medical Systems USA, Inc., Tustin, CA, United States
Synopsis
We prospectively evaluate the generalized performance of the Deep Learning Reconstruction (DLR) method on 55 datasets acquired from 16 different anatomies. For each pulse sequence in each of the 16 anatomies, DLR and 3 predicate methods were reconstructed for randomized blinded review by 3 radiologists based on 8 scoring criteria plus a force-ranking. DLR was scored statistically higher than all 3 predicate methods in 92% of the pairwise comparisons in terms of overall image quality, clinically relevant anatomical/pathological features, and force-ranking. This work demonstrates that DLR generalizes to various anatomies and is frequently preferred over existing methods by experienced readers.
INTRODUCTION
Deep learning reconstruction (DLR) for MR image reconstruction has been translated to clinical practice, where a few DLR solutions have received FDA-510k clearance. The essence of the DLR method is, once trained, to generalize well on the real-world unseen data. This work aims to evaluate the generalized performance of a DLR method [1-3] on data prospectively acquired from multiple anatomies.METHODS
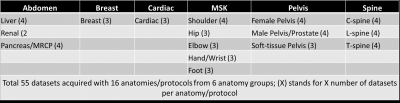
Data acquisition:IRB approval was obtained for the study. Total 55 datasets (from 16 anatomical locations divided into 6 anatomy groups) were acquired from three Canon Orian 1.5T scanners at 3 different imaging sites. Details on the anatomies and the number of datasets per anatomy are listed in Figure 1. There are 5-9 distinct sequences (both 2D and 3D) in each of 16 different protocols resulting in a total of 105 distinct sequences acquired with different orientations and contrast weightings.
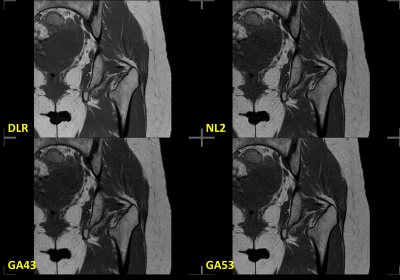
Four different reconstructions (DLR, and other three conventional reconstruction filters: NL2, GA43, and GA53) were performed for each sequence. The labels of the four reconstructions were removed and their order was randomized. The randomized data was shared with 3 ABR-certified radiologists via a cloud-based webPACS for blinded review. The cardiac data was reviewed by 3 separated cardiologists. An example of the 4 reconstructions is shown in Figure 2.
Image Scoring:
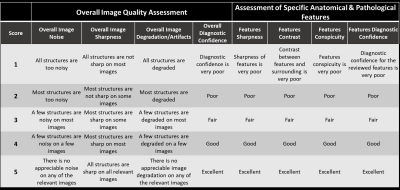
Each of the 4 reconstructions was blindly reviewed and scored based on a Likert scale from 1-5 (1=lowest, 5=highest) on 8 criteria [4] as shown in Figure 3. A key point of the image evaluation is that in addition to the overall image quality assessment (the first 4 criteria), the readers are asked to assess the clinically relevant anatomical and pathological features. Three-to-five clinically relevant anatomical features were pre-specified while pathological features were identified by readers during the review. Additionally, force-ranking from 1-4 (1=worst, 4=best) was performed to assess the readers’ preference among the 4 reconstructions for each sequence.
Analysis:
Scores received from readers were un-blinded and organized for visualization and statistical analysis. Inter-rater agreement was assessed using the Gwet’s Agreement Coefficient (AC2) with ordinal weighting [5, 6]. AC2 values 0–0.20 is indicating as slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1 as almost perfect agreement. Friedman test was used to discern an overall statistically significant difference within the groups, followed by separate Wilcoxon signed-rank tests on the three pairwise comparisons ((i) DLR vs NL2, (ii) DLR vs GA43 and (iii) DLR vs GA53) with a priori Bonferroni adjustment (to correct for the multiple comparisons) for a significance level of 0.017 (i.e., 0.05/3). Data analysis was performed for each of the 6 anatomy groups separately. There are total 162 pairwise comparisons (6 anatomy group x 9 (8 scoring criteria + 1 force-ranking) x 3 pairwise comparisons each comparisons ((i) DLR vs NL2, (ii) DLR vs GA43, and (iii) DLR vs GA53) = 162 total).
RESULTS
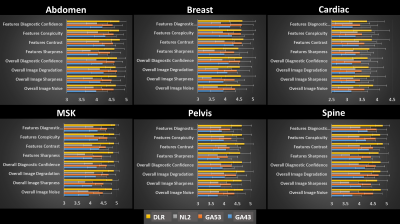
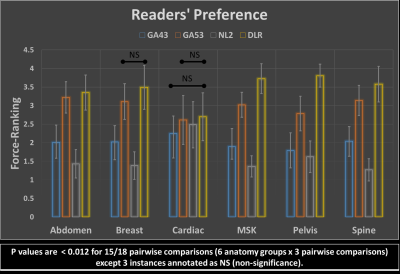
Figures 4 and 5 show averaged scores for each of 8 scoring criteria and the force-ranking scores of 4 reconstructions in all anatomy groups, respectively. DLR’s average score is consistently higher than the other 3 reconstruction methods in 161/162 pairwise comparisons except for one instance where DLR’s average score was smaller than GA53’s by a margin of less than 1%. Gwet’s Agreement Coefficient (AC2) for rater agreement assessment ranges from 0.69 to 0.83, which is considered to be substantial.The Friedman test indicated there were significant differences among the four reconstruction methods. Using the Wilcoxon signed-rank test, significant differences were found within all major anatomy groups and the majority of scoring categories between the three pairwise comparisons. Among all 162 comparisons, there are 149/162 instances where a significant difference was indicated (p<0.017). Specifically for force-ranking, DLR’s average force-ranking score is consistently higher than the other methods in all pairwise comparisons and all anatomy groups. DLR was rated statistically higher (better/preferred) than the other three methods in 15/18 pairwise comparisons (p < 0.012) except the 3 instances annotated as NS (non-significance) in Figure 5.
DISCUSSIONS
This prospective study demonstrates the generalized performance of a Deep Learning Reconstruction method on various anatomies and protocols at 1.5T. Specifically, the DLR is ranked statistically higher in 92% of the pairwise comparisons as compared to the three conventional reconstructions, which are routinely used in clinical practice. For the remainder of the time (8%), there was no difference. Additionally, the force-ranking analysis shows that DLR is preferred over the other reconstruction methods in all studied anatomy groups. It is worth noting that the image scoring was designed to evaluate specific anatomical and pathological features in addition to general and overall image quality assessment. The anatomical features were pre-selected while the pathological features were identified by the readers during the review. This specific design ensures that the clinically relevant anatomical and pathological features were scrutinized and scored to ensure the diagnostic value of the DLR images.CONCLUSION
This blinded multi-reader study demonstrates the generalized performance of the Deep Learning Reconstruction method to the data prospectively acquired on multiple anatomies at 1.5T. Specifically, DLR reconstructed images are preferred by three experts and are scored statistically higher than those reconstructed from the other three routine methods in 92% (149/162) of all pairwise comparisons.Acknowledgements
The authors would like to thank Takeshi Ishimoto, Kensuke Shinoda, Mark Golden, and Shelton Caruthers for their support of this project.References
1. Kidoh M, et al. Deep Learning Based Noise Reduction for Brain MR Imaging: Tests on Phantoms and Healthy Volunteers. Magn Reson Med Sci doi:10.2463/mrms.mp.2019-0018
2. Isogawa K, et al. Noise level adaptive deep convolutional neural network for image denoising. Proceedings of ISMRM, Paris, 2018; 2797.
3. Shinoda K, et al. Deep Learning Based Adaptive Noise Reduction in Multi-Contrast MRI; ISMRM 2019 #4701
4. Cheng JY, Chen F, Sandino C, Mardani M, Pauly JM, Vasanawala SS. Compressed sensing: from research to clinical practice with data-driven learning. arXiv preprint arXiv:1903.07824. 2019 Mar 19.
5. Gwet KL: Handbook of Inter-Rater Reliability. The Definitive Guide to Measuring the Extent of Agreement Among Raters. 2nd edition. Gaithersburg, MD 20886–2696, USA: Advanced Analytics, LLC; 2010.
6. Gwet KL: Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol 2008, 61:29–48.
Figures