1346
Improved multi-shot EPI ghost correction for high gradient strength diffusion MRI using Structured Low-Rank Modeling k-space reconstruction1Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Masachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 2Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 3Q Bio Inc, San Carlos, CA, United States, 4Institute of Medical Physics and Radiation Protection, Mittelhessen University of Applied Sciences, Giessen, Germany, 5Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
Multi-shot EPI diffusion MRI acquired using high diffusion-encoding gradient strengths suffers from severe ghosting artifacts, which can bias and confound the estimation of diffusion microstructural MRI measures at high b-values. In this work, we show that conventional EPI ghost correction techniques fall short in ghosting reduction when high diffusion-encoding gradient strengths ~250mT/m are used, and that advanced reconstruction algorithms based on structured low-rank matrix modeling can substantially reduce ghosting without introducing additional artifacts.
Introduction
Multi-shot EPI diffusion MRI is an appealing alternative to conventional single-shot EPI dMRI as it is less vulnerable to geometric distortions and eddy-current artifacts due to the shortened echo train length1. Unfortunately, reconstruction of multi-shot EPI images still suffers from odd/even line inconsistency, an intrinsic feature to all EPI sequences2 (i.e., Nyquist ghosting). These effects are exacerbated when using high gradient strength MR scanners as the strong gradients amplify eddy current distortions and static field inhomogeneities3.To address this problem, we explore the use of advanced reconstruction algorithms based on Structured-Low Rank matrix (SLM) modeling4-7, which have shown excellent results for 2D single and multi-shot EPI dMRI with lower diffusion gradient strengths. We demonstrate that SLM modeling can provide high quality, artifact-free 3D multi-shot dMRI EPI with diffusion gradient strengths up to 250 mT/m.
Method
Ghosting is a pervasive artifact in EPI, which is produced by the mismatches between k-space data acquired with gradients with different polarities8 and multiple shots. Conventionally, EPI ghosting is corrected with phase information from k-space data acquired from navigator scans. While robust and computationally cheap, ghost correction methods based on navigators often make modeling errors. The assumptions made in correcting for k-space line mismatches are excessively simplistic, especially in the regime of high gradient strengths.Alternatively, it has been shown that ghost correction in EPI reconstruction can be cast as an undersampled k-space reconstruction problem, where the k-space data of different shots and polarities are treated separately. As the k-space data for each shot and polarity is highly undersampled, accurate reconstruction requires prior knowledge to compensate for the lack of measured data (ill-posed problem). Structured Low-rank Matrix (SLM) methods exploit the fact that k-space data are linearly predictable, and hence it can be embedded into a structured Hankel matrix, which can be shown to be a low-rank matrix9. Hence, missing k-space lines can be recovered by applying low-rank matrix recovery techniques.
We investigate whether SLM methods can reduce ghosting artifacts in ex-vivo whole human brain 3D diffusion EPI data acquired with high diffusion-encoding gradient. Similar to the work of 10, we formulate the reconstruction problem as a SENSE-type optimization algorithm, where the data fidelity term is complemented with a regularizer based on SLM. This regularization term enforces low-rank characteristics on a Hankel matrix that comprises of the k-space data from all shots and odd/even polarities. The proposed method can be considered within the LORAKS framework4, and we refer to it as the LORAKS-based method from now on. Coil sensitivities are estimated from a GRE acquisition with the popular ESPIRiT algorithm11. We compare this approach with the standard linear phase correction (LPC) method based on 1D navigators8, and with a SENSE-based reconstruction with no additional prior information
Data:
All data were acquired on a dedicated high-gradient 3T MRI scanner (MAGNETOM Connectom, Siemens Healthineers) capable of maximum gradient strength of 300 mT/m. A custom-made, 48-channel phased array coil for ex vivo whole human brain imaging was used for signal reception12. Ex-vivo whole-brain diffusion MR data were acquired at 1 mm isotropic resolution with a Stejskal-Tanner, 3D multi-shot EPI sequence. Relevant acquisition parameters were: matrix size = 196 x 124 x 160, 4 shots, EPI factor = 31, no partial Fourier, head-foot phase-encoding direction, TE/TR = 53/500 ms, and maximum gradient strength of 253 mT/m. The diffusion MRI protocol consisted of one b=0 image and 16 diffusion-weighted volumes at b=4,000 s/mm2 encoded with diffusion directions that were spread over the sphere.
Algorithm implementation and post-processing:
The LORAKS-based method was implemented in a 2D fashion. The 3D k-space data (kx x ky x kz) was inverse-Fourier transformed along kz, and three representatives slices z, were extracted from the hybrid (kx x ky x z) data set. The FISTA algorithm was used for optimization10. Reconstructed images for all shots and polarities were combined with the root sum of squares (rSoS) technique, as was also for LPC and SENSE methods.
Results
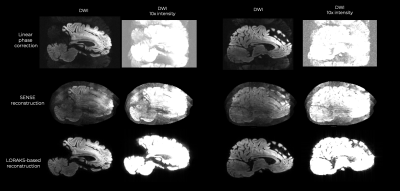
Reconstructed images using the combined shots are shown in Fig.1. The LPC and SENSE-based reconstructions were insufficient to correct for the ghosting artifacts seen with high diffusion-encoding gradient strengths. On the other hand, LORAKS enabled substantial ghost reduction, which is best seen on the intensity-amplified images.Discussion
Our results show that EPI ghosting in images acquired at high diffusion-encoding gradient strengths requires more complex approaches than conventional 1D navigators, and that SLM-based recovery algorithms can substantially reduce the level of ghosting. We foresee an even better quality reconstruction with more advanced SLM-algorithms i.e., exploiting linear predictability of k-space data over channels5, introducing calibration data into the reconstruction problem13, or incorporating q-space regularizers, aiming at a joint reconstruction from the k-q space, directly14-16.Conclusions
3D multi-shot diffusion MRI acquired using high diffusion-encoding gradients requires far more advanced reconstruction algorithms than conventional approaches to producing ghosting-free DWI images. In this preliminary work, we have shown that structured low-rank matrix-based recovery algorithms substantially reduce ghosting effects, enabling distortion-free, diffusion MRI in ex vivo samples that often require higher b-values to achieve sufficient diffusion weighting. We believe SLM-based algorithms could be indispensable for high quality, multi-shot diffusion MRI reconstruction in Connectome like MR scanners.Acknowledgements
This work was supported by the NIH (grants P41-EB030006, U01-EB026996, R01-EB021265, K23-NS096056).References
1. Holdsworth, S. J. et al. The quest for high spatial resolution diffusion‐weighted imaging of the human brain in vivo. NMR in Biomedicine, 2019, 32(4), e40562.
2. Schmitt, F. et al. Echo-planar imaging: theory, technique, and application. Springer Science & Business Media.
3. Setsompop, K et al. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage, 80, 220-233.
4. Haldar, J. P. Low-rank modeling of local k-space neighborhoods (LORAKS) for constrained MRI. IEEE Transactions on Medical Imaging, 33(3), 668-681.
5. Lobos, R. A. et al. Navigator-free EPI ghost correction with structured low-rank matrix models: New theory and methods. IEEE Transactions on Medical Imaging, 37(11), 2390-2402.
6. Mani, Merry, et al. "Multi‐shot sensitivity‐encoded diffusion data recovery using structured low‐rank matrix completion (MUSSELS)." Magnetic resonance in medicine 78(2): 494-507.
7. Lee, Juyoun et al. "Reference‐free single‐pass EPI Nyquist ghost correction using annihilating filter‐based low rank Hankel matrix (ALOHA)." Magnetic resonance in medicine 76(6): 1775-1789.
8. Buonocore, M. H., & Gao, L.. Ghost artifact reduction for echo-planar imaging using image phase correction. Magnetic resonance in medicine, 38(1), 89-100.
9. Haldar, J. P., & Setsompop, K. Linear predictability in MRI reconstruction: Leveraging shift-invariant Fourier structure for faster and better imaging. IEEE Signal Process. Mag., 37(1) 69-82
10. Bilgic, Berkin, et al. "Highly accelerated multishot echo-planar imaging through synergistic machine learning and joint reconstruction." Magnetic resonance in medicine 82(4): 1343-1358.
11. Uecker, Martin, et al. "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA." Magnetic resonance in medicine 71(3): 990-1001.
12. Scholz, A, et al., “A 48-channel ex vivo Brain Array Coil for Diffusion-Weighted MRI at 3T”, Proc. Intl. Soc. Mag. Reson. Med. 27, 1494
13. Lobos, R. A. et al., "Robust Autocalibrated Structured Low-Rank EPI Ghost Correction", Magnetic resonance in Medicine, 2020;00:1-17
14. Hu, Y. et al.. Multi‐shot diffusion‐weighted MRI reconstruction with magnitude‐based spatial‐angular locally low‐rank regularization (SPA‐LLR). Magnetic Resonance in Medicine, 83(5), 1596-1607.
15. Ramos‐Llordén, G. et al. High‐fidelity, accelerated whole‐brain submillimeter in vivo diffusion MRI using gSlider‐spherical ridgelets (gSlider‐SR). Magnetic Resonance in Medicine., 84(4) 1781-1795
16. Wu, W. et al. Diffusion acceleration with Gaussian process estimated reconstruction (DAGER). Magnetic resonance in medicine, 82(1), 107-125.